How to read your child’s genetic test results
By Ji-Sun Kim, MSc, MS, CGC
Certified Genetic Counselor
Weill Cornell Medicine
Center for Neurogenetics
When you receive a diagnosis of MEF2C haploinsufficiency syndrome, you likely underwent genetic testing. There are a few different types of genetic tests that could have led to a diagnosis: a chromosomal microarray, epilepsy gene panel, or whole exome or whole genome sequencing. For each type of testing, there could be different “flavors” of results that could come up.
Let’s take a step back and think about what we mean by genes and genetic testing. If you think of our genome as a book, then chromosomal microdeletion could be missing a chapter. Sequencing changes could be spell changes, or missing words. In the same vein, a mutation could be like a missing a letter in a word, or a letter switched to another letter. We often use the terms “mutation” and “variant” interchangeably.
From Greenwood Genetics
A chromosomal microarray checks for chromosomal material that is in the cells—it is checking for missing or extra pieces of material. In our book analogy, that would be like missing a chapter or a few chapters. If there are missing pieces of genome, then the result could say “deletion” near the location of the MEF2C gene, which is at 5q14.3.—at chromosome 5, long arm Q, at location 14.3. Typically, this would also include the size of the deletion with a number. The size of the MEF2C gene is around 200kb (200,000 base pairs), the deleted part could be larger than this area, or sometimes smaller than 200kb. The gene is sectioned into exons, which code for the amino acids, which code for the protein. Three exons code for the “MADS” domain, which is critical for the normal functioning of the MEF2C protein. This domain typically is up to amino acid number 60, as depicted in the below diagram. Genetic testing reports will not necessarily tell you which exon your mutation is located; some do, but not all. The exact location of your child’s mutation on the MEF2C gene is a question you could ask your provider, or could look up yourself.
Chen X, Gao B, Ponnusamy M, Lin Z, Liu J. MEF2 signaling and human diseases. Oncotarget. 2017 Dec;8(67):112152-112165. DOI: 10.18632/oncotarget.22899. PMID: 29340119; PMCID: PMC5762387.
Sometimes, a genetic report will indicate the genomic coordinate of your child’s mutation, and this may look something like chr5:88100017-88379216 del or chr5: (88100017-88379216)x1. X1 indicates that instead of having two copies of the coordinate, which would be the typical situation, as we would inherit one copy from mom, and another copy from dad, there is only one copy present, hence a deletion.
A genetic testing report is likely to read something like this:
arr 5q14.3 (87, 516,643 – 90, 382, 981) x1
arr= the analysis was by microarray
5q14.3= the analysis revealed a change in band 5q14.3
(87, 516,643 – 90, 382, 981)x1 = the specific DNA base pairs that are missing. The first base pair shown to be missing is at location 87, 516, 643 counting from the top of the chromosome, and the last base pair missing at 90, 382, 981. The difference of the two locations would indicate the size of the deletion, in this case, could be written as 2,866, 338 base pairs, or in short, 2.9Mb.
If you have had epilepsy gene panel or whole exome/genome sequencing, the report would have different language, often with letters and numbers. This is usually organized with c. location identifier, p. location identifier, followed by the gene name, and I am assuming it would be MEF2C. For example, it would look something like c.123 A>G, p. Leu35Pro, MEF2C. The report may include variants noted in other genes as well.
The c., or “C-dot” indicates where along the DNA sequence the change, or the “variant” was observed. The “c” is a code for cDNA, which is the part of the DNA that gets converted to mRNA, which ultimately goes on to make the protein. Proteins are composed of a sequence of amino acids. The c. number tells us the exact location of the change along the DNA sequence of the MEF2C gene. DNA is coded by 4 different nucleotides, A, C, G, T. the report will indicate what exactly the switch is. For example, c.123 A>G means at the 123th location along the cDNA, it is typically nucleotide A, but is switched to G instead. Sometimes, the report would indicate the genomic coordinate instead of cDNA, and this may look something like chr5:88100017 G-A. Again, this means that at the 8810017th location of the genomic DNA, the G nucleotide is switched to A.
The p., or “P-dot” tells us the protein level change as the consequence of the DNA change. For example, p. Leu35Pro means that at the 35th location, it is usually amino acid Leucine, but because of the DNA change, it is now switched to Proline, a different amino acid with different properties. This type of change can inherently impact the normal shape, thus the intended functioning of the protein. Sometimes the cDNA change could lead to a stop, indicated by *. An example is p. Arg207*. This means that at 207th location codes for Arginine, but now the protein stops there.
Sometimes, there may be missing or deletions at the nucleotide level, and this could be indicated as “del” or addition of nucleotides, indicated as “dup” (if the same existing nucleotides) or “ins” if different nucleotides are inserted.
Here is another example: c.204_208del, p. Lys68Asnfs*8
In this example, there was a few nucleotides that were deleted from 204 to 208 location, and this would lead to not only the switch from lysine to asparagine at 68th location of the amino acid, also a premature stop codon is introduced 8 amino acids downstream from the frameshift. This ultimately truncates the protein.
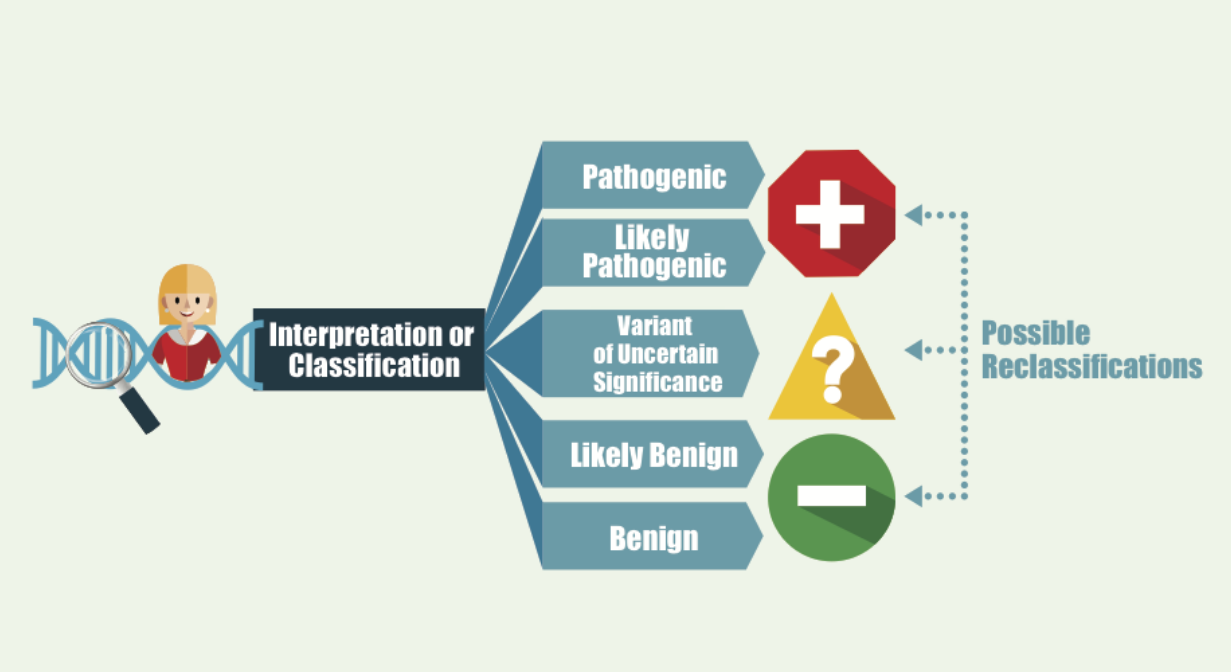
Once the genetic variant is identified – whether a deletion, or a sequence change, then the lab will report out the classification of the that. Think of this was a spectrum, not black and white type of answer. For each variant, it is “graded” based on many factors, the resulting classification is reported, and this is bit of a spectrum. With the strongest evidence available – the lab would say a variant is “pathogenic”, meaning with a very high confidence level that this genetic change is disease-causing. Then level down, is likely pathogenic – the lab is fairly certain that the genetic change is leading to the disease but may lack some functional study or animal study information for that specific variant. Doctors or clinicians usually treat pathogenic and likely pathogenic variants similarly. This would confirm the genetic diagnosis in your child.
The next level of classification is called variant of uncertain significance, or VUS. This means that the lab observes a genetic variant but is not 100% confident that this change is leading to a disease in the patient. For neurodevelopmental conditions like MEF2C haploinsufficiency, the de novo nature of the mutation (that is, not inherited from either parent) plays a significant role in the mutation being classified as disease causing. Thus, if the genetic testing was only done on the child as a gene panel or proband only whole exome, then the lab may classify the variant as VUS. Once parental samples are submitted, and confirm that it is a de novo variant, then the lab may reclassify the variant as likely pathogenic. Also, if the genetic variant has never been observed previously in anyone else, then the lab may not have the highest level of confidence to classify as disease-causing. They may wait to see it in other affected individuals before reclassifying. Of note, the variant classification of pathogenic vs. likely pathogenic vs. VUS does not mean or correlate to the severity of the child’s symptom. Within each classification, there could be a wide range of phenotypic spectrum.
The goal of the Volare Study at Weill Cornell is to better understand the genotype-phenotype correlation of the MEF2C syndrome, i.e. if a variant is correlated with specific types of clinical picture, whether it is severe vs. mild.
MEF2C gene has 12 exons in total. Exons are ultimately transcribed and translated into protein. It is believed that the first 3 exons are critical for the normal functioning of the protein, and any variations in this region is deemed highly sensitive, i.e. disease causing. We are suspecting that variants in this region may also lead to a more severe clinical picture, while variants found elsewhere along the gene if noted towards the end of the gene could lead to a milder symptom. We are hoping to gather more information regarding this through our study!
The majority of the reported MEF2C cases occur de novo, meaning the variation is not being passed down from a parent. The mutation most likely occurred during egg or sperm formation or shortly after embryo formation with error-free egg and sperm that fertilized. There is really no way of knowing. But because of this, the likelihood of this happening in subsequent pregnancies is low, around 1%. In rare cases, germline mosaicism – some of the sperm cells or egg cells having the mutation, even if the parent of origin is healthy – has been reported. We tell parents that it is unlikely to happen again in subsequent pregnancies, but once a mutation is identified in your child, then it is possible to check in your subsequent pregnancies. Some parents elect to undergo IVF pregnancies and check the embryos prior to implanting.